Ubiquitin D
Ubiquitin D is a protein that in humans is encoded by the UBD gene, also known as FAT10.[5][6][7] UBD acts like ubiquitin, by covalently modifying proteins and tagging them for destruction in the proteasome.
Ubiquitin
[edit]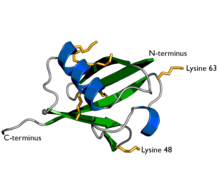
Ubiquitin is a protein composed of 76 amino acids. In order for ubiquitin to bind to other proteins, it must go through an activation process by E1, an ATP-dependent ubiquitin activating enzyme. The carboxyl terminal (C-terminus) of ubiquitin is linked to the cysteine residue of the E1 protein by a high energy thioester linkage and activated. This reaction requires ATP and proceeds through a covalent AMP-ubiquitin intermediate.
Importance of Ubiquitin
[edit]Proteolysis in cells is an essential process to prevent the production of unwanted or abnormal proteins. During protein degradation, the process in which proteolytic enzymes act in an ATP-dependent manner mainly occurs in the degradation of substrate proteins with short half-lives. In particular, in eukaryotic cells, the proteolysis process by the binding of ubiquitin composed of 76 amino acids plays an important role in regulating the half-life and function of proteins.[8]
Ubiquitin-binding E1, E2, and E3 enzymes
[edit]E1, E2 proteins and E3 enzymes are essential for attaching ubiquitin to matrix proteins. Among them, only one type of E1 protein exists in an individual, and it is known that the number is the highest.
About a dozen E2 proteins are known to exist in yeast as transfer proteins that transfer ubiquitin from E1 to E3 or substrates, and it has been confirmed that more types exist in higher eukaryotes than yeast. E3 enzyme, also called E3 ligase, is an enzyme that acts in the final step of attaching ubiquitin to a substrate protein. The specificity of the substrate protein to be ubiquitinated is determined by the E3 enzyme.
In particular, E3 enzymes can be divided into four types such as HECT, RING-finger, U-box, and PHD-finger according to the method of determining the specificity of the substrate protein. Among them, the RING-finger E3 enzyme is known to follow the amino terminal rule the best. The amino-terminal rule means that the degradation rate of a protein, that is, the half-life of a protein, varies depending on what the amino-terminal amino acid residue is. In the amino-terminal rule of yeast, it is known that the half-life of proteins varies depending on the presence of 12 unstable amino acid residues (arginine, lysine, histidine, tyrosine, tryptophan, isoleucine, aspartic acid, glutamic acid, asparagine, glutamine) out of 20 amino acids. When methionine, glycine, serine, valine, or proline is located at the amino-terminus, it has a half-life of more than 20 hours, whereas when residues such as tyrosine, glutamine, leucine, phenylalanine, aspartate, lysine, or arginine are located, the half-life is reduced to less than 10 minutes.[9]
Activated ubiquitin bound to E1 is transferred to the cysteine residue of the E2 molecule (ubiquitin conjugating enzyme). E2 molecules form complexes with E3 protein molecules, which are accessory proteins. In the E2-E3 complex, called ubiquitin ligase, the E3 component binds to a specific degradation signal in the substrate protein, and then the E2 enzyme helps to form a multi-ubiquitin chain at the lysine residue of the substrate protein. In order to form a multi-ubiquitin chain, the C-terminal region of the next ubiquitin molecule is linked to the lysine residue of the preceding ubiquitin.
About 30 different E2s exist in mammalian cells, and more than 300 specific E2-E3 complexes have been found. Thanks to the function of the E3 component, they can recognize specific degradation signals of target proteins. In general, E2 is called ubiquitin-linking enzyme, and E3 is traditionally called ubiquitin-linking enzyme. Functionally, however, it is more accurate to call the E2-E3 complex a ubiquitin ligase. Multi-ubiquitin chains on target proteins are recognized by specific receptors on the proteasome.
In general, the proteasome is responsible for removing structurally abnormal proteins, but also contributes to regulating the lifespan of specific proteins according to the intracellular environment. For example, mitotic cyclins required for the induction of mitosis (or mitosis) remain long-lived throughout the cell cycle, but are destined for abrupt degradation at the end of mitosis.
Then, how does this regulation of protein degradation occur? Activation of E3, a ubiquitin linking enzyme, can be achieved by phosphorylation or allosteric transition following ligand or subunit binding. For example, the anaphase-promoting complex (APC) is a multiubiquitin-linking enzyme that is activated during mitosis by the addition of subunits along the cell cycle. Activated APC promotes proteolysis of mitotic cyclins and metaphase-late transition regulators.[10]
UBD
[edit]UBD was first discovered in reticuloendothelial tissues and mucosal-associated lymphoid immunological systems as one of the genes at the human major histocompatibility complex class I locus on chromosome 6. The UBD gene encodes an 18 kDa protein containing an N- and C-terminus with 29 and 36% identity with ubiquitin, respectively. Of the ubiquitin-like proteins that have been identified, UBD is the only one of that conjugate to target proteins by a free diglycine motif at the C-terminus and directly guides noncovalently bound proteins to proteasomal degradation.
UBD also has important roles in cell mitosis, chromosome instability, apoptosis and immune response. UBD deregulation may induce abnormal alterations in apoptosis, cell division or chromosome instability, which are associated with neoplastic change. Tumor UBD expression shows some tissue specificity, with transcriptional upregulation observed in liver, uterine cervix, ovarian, pancreatic, gastric and small intestine adenocarcinomas, but not in thyroid, prostate or kidney cancers. In hepatic cancer cells, increased expression of UBD was associated with Proliferating Cell Nuclear Antigen, a cell proliferation marker, and reported to provide a growth advantage over cells without UBD expression. High UBD expression also promoted hepatocellular carcinoma development in a mouse model and formation of Mallory–Denk bodies, which are preneoplastic changes in chronic liver disease. Overexpression of UBD in gastric cancer has been correlated with metastasis and tumor staging, and both UBD mRNA and protein levels were identified as independent prognostic factors for this disease. Increased UBD has also been positively correlated with mutant p53 expression, which may activate UBD expression and indirectly facilitate gastric cancer progression. Interferon-γ and tumor necrosis factor-α act synergistically to induce the UBD promoter through an interferon sequence resposive element. Collectively, these data indicate that UBD may be a marker for precancerous lesions and may promote cancer progression.[11]
Interactions
[edit]UBD has been shown to interact with NUB1[12] and MAD2L1.[13]
References
[edit]- ^ a b c ENSG00000231968, ENSG00000206468, ENSG00000206513, ENSG00000224654, ENSG00000213886, ENSG00000226898 GRCh38: Ensembl release 89: ENSG00000228913, ENSG00000231968, ENSG00000206468, ENSG00000206513, ENSG00000224654, ENSG00000213886, ENSG00000226898 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000035186 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Bates EE, Ravel O, Dieu MC, Ho S, Guret C, Bridon JM, Ait-Yahia S, Brière F, Caux C, Banchereau J, Lebecque S (Oct 1997). "Identification and analysis of a novel member of the ubiquitin family expressed in dendritic cells and mature B cells". European Journal of Immunology. 27 (10): 2471–7. doi:10.1002/eji.1830271002. PMID 9368598. S2CID 21652482.
- ^ Fan W, Cai W, Parimoo S, Schwarz DC, Lennon GG, Weissman SM (Aug 1996). "Identification of seven new human MHC class I region genes around the HLA-F locus". Immunogenetics. 44 (2): 97–103. doi:10.1007/BF02660056. PMID 8662070. S2CID 21628804.
- ^ "Entrez Gene: UBD ubiquitin D".
- ^ "유비퀴틴". terms.naver.com (in Korean). Retrieved 2023-04-27.
- ^ "유비퀴틴". terms.naver.com (in Korean). Retrieved 2023-04-27.
- ^ "유비퀴틴". terms.naver.com (in Korean). Retrieved 2023-04-27.
- ^ Yan DW, Li DW, Yang YX, Xia J, Wang XL, Zhou CZ, Fan JW, Wen YG, Sun HC, Wang Q, Qiu GQ, Tang HM, Peng ZH (September 2010). "Ubiquitin D is correlated with colon cancer progression and predicts recurrence for stage II-III disease after curative surgery". British Journal of Cancer. 103 (7): 961–969. doi:10.1038/sj.bjc.6605870. ISSN 1532-1827. PMC 2965875. PMID 20808312.
- ^ Hipp MS, Raasi S, Groettrup M, Schmidtke G (Apr 2004). "NEDD8 ultimate buster-1L interacts with the ubiquitin-like protein FAT10 and accelerates its degradation". The Journal of Biological Chemistry. 279 (16): 16503–10. doi:10.1074/jbc.M310114200. PMID 14757770.
- ^ Liu YC, Pan J, Zhang C, Fan W, Collinge M, Bender JR, Weissman SM (Apr 1999). "A MHC-encoded ubiquitin-like protein (FAT10) binds noncovalently to the spindle assembly checkpoint protein MAD2". Proceedings of the National Academy of Sciences of the United States of America. 96 (8): 4313–8. Bibcode:1999PNAS...96.4313L. doi:10.1073/pnas.96.8.4313. PMC 16329. PMID 10200259.
Further reading
[edit]- Mazzé FM, Degrève L (2006). "The role of viral and cellular proteins in the budding of human immunodeficiency virus". Acta Virologica. 50 (2): 75–85. PMID 16808324.
- Ott DE, Coren LV, Copeland TD, Kane BP, Johnson DG, Sowder RC, Yoshinaka Y, Oroszlan S, Arthur LO, Henderson LE (Apr 1998). "Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and to the p12Gag protein of Moloney murine leukemia virus". Journal of Virology. 72 (4): 2962–8. doi:10.1128/JVI.72.4.2962-2968.1998. PMC 109742. PMID 9525617.
- Liu YC, Pan J, Zhang C, Fan W, Collinge M, Bender JR, Weissman SM (Apr 1999). "A MHC-encoded ubiquitin-like protein (FAT10) binds noncovalently to the spindle assembly checkpoint protein MAD2". Proceedings of the National Academy of Sciences of the United States of America. 96 (8): 4313–8. Bibcode:1999PNAS...96.4313L. doi:10.1073/pnas.96.8.4313. PMC 16329. PMID 10200259.
- Schubert U, Ott DE, Chertova EN, Welker R, Tessmer U, Princiotta MF, Bennink JR, Krausslich HG, Yewdell JW (Nov 2000). "Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of HIV-1 and HIV-2". Proceedings of the National Academy of Sciences of the United States of America. 97 (24): 13057–62. Bibcode:2000PNAS...9713057S. doi:10.1073/pnas.97.24.13057. PMC 27177. PMID 11087859.
- Strack B, Calistri A, Accola MA, Palu G, Gottlinger HG (Nov 2000). "A role for ubiquitin ligase recruitment in retrovirus release". Proceedings of the National Academy of Sciences of the United States of America. 97 (24): 13063–8. Bibcode:2000PNAS...9713063S. doi:10.1073/pnas.97.24.13063. PMC 27178. PMID 11087860.
- Ott DE, Coren LV, Chertova EN, Gagliardi TD, Schubert U (Dec 2000). "Ubiquitination of HIV-1 and MuLV Gag". Virology. 278 (1): 111–21. doi:10.1006/viro.2000.0648. PMID 11112487.
- Strack B, Calistri A, Göttlinger HG (Jun 2002). "Late assembly domain function can exhibit context dependence and involves ubiquitin residues implicated in endocytosis". Journal of Virology. 76 (11): 5472–9. doi:10.1128/JVI.76.11.5472-5479.2002. PMC 137019. PMID 11991975.
- Ott DE, Coren LV, Sowder RC, Adams J, Schubert U (Mar 2003). "Retroviruses have differing requirements for proteasome function in the budding process". Journal of Virology. 77 (6): 3384–93. doi:10.1128/JVI.77.6.3384-3393.2003. PMC 149504. PMID 12610113.
- Lee CG, Ren J, Cheong IS, Ban KH, Ooi LL, Yong Tan S, Kan A, Nuchprayoon I, Jin R, Lee KH, Choti M, Lee LA (May 2003). "Expression of the FAT10 gene is highly upregulated in hepatocellular carcinoma and other gastrointestinal and gynecological cancers". Oncogene. 22 (17): 2592–603. doi:10.1038/sj.onc.1206337. PMID 12730673.
- Brès V, Kiernan RE, Linares LK, Chable-Bessia C, Plechakova O, Tréand C, Emiliani S, Peloponese JM, Jeang KT, Coux O, Scheffner M, Benkirane M (Aug 2003). "A non-proteolytic role for ubiquitin in Tat-mediated transactivation of the HIV-1 promoter". Nature Cell Biology. 5 (8): 754–61. doi:10.1038/ncb1023. PMID 12883554. S2CID 8414608.
- Hipp MS, Raasi S, Groettrup M, Schmidtke G (Apr 2004). "NEDD8 ultimate buster-1L interacts with the ubiquitin-like protein FAT10 and accelerates its degradation". The Journal of Biological Chemistry. 279 (16): 16503–10. doi:10.1074/jbc.M310114200. PMID 14757770.
- Barrios-Rodiles M, Brown KR, Ozdamar B, Bose R, Liu Z, Donovan RS, Shinjo F, Liu Y, Dembowy J, Taylor IW, Luga V, Przulj N, Robinson M, Suzuki H, Hayashizaki Y, Jurisica I, Wrana JL (Mar 2005). "High-throughput mapping of a dynamic signaling network in mammalian cells". Science. 307 (5715): 1621–5. Bibcode:2005Sci...307.1621B. doi:10.1126/science.1105776. PMID 15761153. S2CID 39457788.
- Hipp MS, Kalveram B, Raasi S, Groettrup M, Schmidtke G (May 2005). "FAT10, a ubiquitin-independent signal for proteasomal degradation". Molecular and Cellular Biology. 25 (9): 3483–91. doi:10.1128/MCB.25.9.3483-3491.2005. PMC 1084302. PMID 15831455.
- Gottwein E, Kräusslich HG (Jul 2005). "Analysis of human immunodeficiency virus type 1 Gag ubiquitination". Journal of Virology. 79 (14): 9134–44. doi:10.1128/JVI.79.14.9134-9144.2005. PMC 1168789. PMID 15994808.
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M (Oct 2005). "Towards a proteome-scale map of the human protein-protein interaction network". Nature. 437 (7062): 1173–8. Bibcode:2005Natur.437.1173R. doi:10.1038/nature04209. PMID 16189514. S2CID 4427026.




